Full matrix models¶
For specific reference measures the \(\beta = 1, 2, 4\) cases are very singular in the sense that the corresponding ensembles coincide with the eigenvalues of random matrices.
This is a highway for sampling exactly such ensembles in \(\mathcal{O}(N^3)\)!
Hermite Ensemble¶
Take for reference measure \(\mu=\mathcal{N}(0, 2)\), the pdf of the corresponding \(\beta\)-Ensemble reads
Hint
The Hermite ensemble (whose name comes from the fact that Hermite polynomials are orthogonal w.r.t the Gaussian distribution) refers to the eigenvalue distribution of random matrices formed by i.i.d. Gaussian vectors.
\(\beta=1\)
\(\beta=2\)
\(\beta=4\)
Normalization \(\sqrt{\beta N}\) to concentrate as the semi-circle law.
from dppy.beta_ensembles import HermiteEnsemble
hermite = HermiteEnsemble(beta=4) # beta in {0,1,2,4}, default beta=2
hermite.sample_full_model(size_N=500)
# hermite.plot(normalization=True)
hermite.hist(normalization=True)
# To compare with the sampling speed of the tridiagonal model simply use
# hermite.sample_banded_model(size_N=500)
(Source code, png, hires.png, pdf)
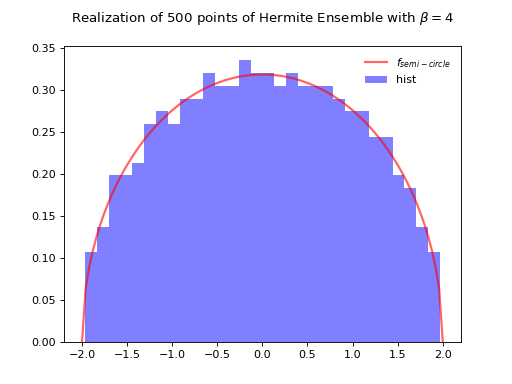
Fig. 8 Full matrix model for the Hermite ensemble¶
See also
Banded matrix model for Hermite ensemble
HermiteEnsemblein API
Laguerre Ensemble¶
Take for reference measure \(\mu=\Gamma\left(\frac{\beta}{2}(M-N+1), 2\right)=\chi_{\beta(M-N+1)}^2\), the pdf of the corresponding \(\beta\)-Ensemble reads
Hint
The Laguerre ensemble (whose name comes from the fact that Laguerre polynomials are orthogonal w.r.t the Gamma distribution) refers to the eigenvalue distribution of empirical covariance matrices of i.i.d. Gaussian vectors.
\(\beta=1\)
\(\beta=2\)
\(\beta=4\)
Normalization \(\beta M\) to concentrate as Marcenko-Pastur law.
where
from dppy.beta_ensembles import LaguerreEnsemble
laguerre = LaguerreEnsemble(beta=1) # beta in {0,1,2,4}, default beta=2
laguerre.sample_full_model(size_N=500, size_M=800) # M >= N
# laguerre.plot(normalization=True)
laguerre.hist(normalization=True)
# To compare with the sampling speed of the tridiagonal model simply use
# laguerre.sample_banded_model(size_N=500, size_M=800)
(Source code, png, hires.png, pdf)
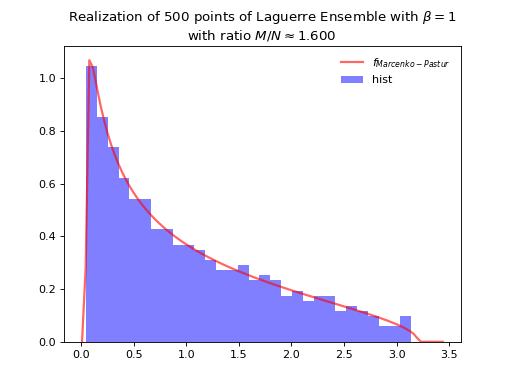
Fig. 10 Full matrix model for the Laguerre ensemble¶
See also
Banded matrix model for Laguerre ensemble
LaguerreEnsemblein API
Jacobi Ensemble¶
Take for reference measure \(\mu=\operatorname{Beta}\left(\frac{\beta}{2}(M_1-N+1), \frac{\beta}{2}(M_2-N+1)\right)\), the pdf of the corresponding \(\beta\)-Ensemble reads
Hint
The Jacobi ensemble (whose name comes from the fact that Jacobi polynomials are orthogonal w.r.t the Beta distribution) is associated with the multivariate analysis of variance (MANOVA) model.
\(\beta=1\)
\(\beta=2\)
\(\beta=4\)
Concentrates as Wachter law
where
itself tending to the arcsine law in the limit.
from dppy.beta_ensembles import JacobiEnsemble
jacobi = JacobiEnsemble(beta=2) # beta must be in {0,1,2,4}, default beta=2
jacobi.sample_full_model(size_N=400, size_M1=500, size_M2=600) # M_1, M_2 >= N
# jacobi.plot(normalization=True)
jacobi.hist(normalization=True)
# To compare with the sampling speed of the triadiagonal model simply use
# jacobi.sample_banded_model(size_N=400, size_M1=500, size_M2=600)
(Source code, png, hires.png, pdf)
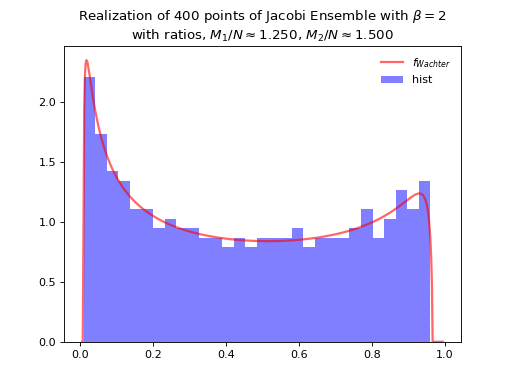
Fig. 12 Full matrix model for the Jacobi ensemble¶
See also
Banded matrix model for Jacobi ensemble
JacobiEnsemblein APIMultivariateJacobiOPEin API
Circular Ensemble¶
Hint
Eigenvalues of orthogonal (resp. unitary and self-dual unitary) matrices drawn uniformly i.e. Haar measure on the respective groups. The eigenvalues lie on the unit circle i.e. \(\lambda_n = e^{i \theta_n}\). The distribution of the angles \(\theta_n\) converges to the uniform measure on \([0, 2\pi[\) as \(N\) grows.
\(\beta=1\)
Uniform measure i.e. Haar measure on orthogonal matrices \(\mathbb{O}_N\): \(U^{\top}U = I_N\)
Via QR algorithm, see [Mez06] Section 5
import numpy as np from numpy.random import randn import scipy.linalg as la A = randn(N, N) Q, R = la.qr(A) d = np.diagonal(R) U = np.multiply(Q, d/np.abs(d), Q) la.eigvals(U)
The Hermite way
\[\begin{split} X \sim \mathcal{N}_{N, N}(0,1)\\ A = X+X^{\top} = U^{\top}\Lambda U\\ eigvals(U).\end{split}\]\(\beta=2\)
Uniform measure i.e. Haar measure on unitary matrices \(\mathbb{U}_N\): \(U^{\dagger}U = I_N\)
Via QR algorithm, see [Mez06] Section 5
import numpy as np from numpy.random import randn import scipy.linalg as la A = randn(N, N) + 1j*randn(N, N) Q, R = la.qr(A) d = np.diagonal(R) U = np.multiply(Q, d / np.abs(d), Q) la.eigvals(U)
from dppy.beta_ensembles import CircularEnsemble circular = CircularEnsemble(beta=2) # beta in {0,1,2,4}, default beta=2 # 1. Plot the eigenvalues, they lie on the unit circle circular.sample_full_model(size_N=30, haar_mode='QR') circular.plot() # 2. Histogram of the angle of more points, should look uniform on [0,2pi] circular.flush_samples() # Flush previous sample circular.sample_full_model(size_N=1000, haar_mode='QR') circular.hist()
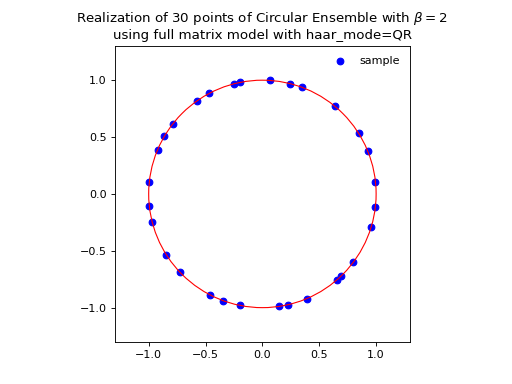
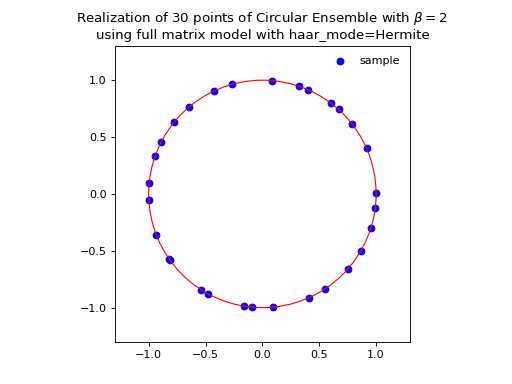
Fig. 14 (png, hires.png, pdf) Full matrix model for the Circular ensemble from QR on random Gaussian matrix¶
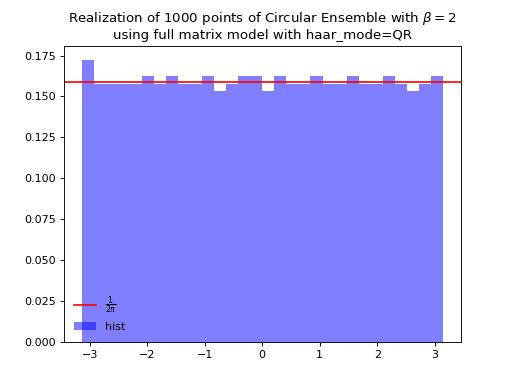
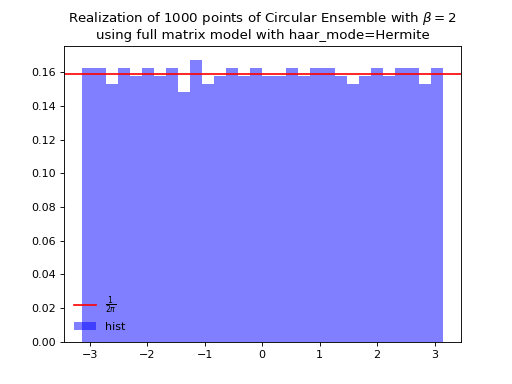
Fig. 15 (png, hires.png, pdf) Full matrix model for the Circular ensemble from QR on random Gaussian matrix¶
The Hermite way
\[\begin{split} X \sim \mathcal{N}_{N, N}(0,1) + i~ \mathcal{N}_{N, N}(0,1)\\ A = X+X^{\dagger} = U^{\dagger}\Lambda U\\ eigvals(U).\end{split}\]from dppy.beta_ensembles import CircularEnsemble circular = CircularEnsemble(beta=2) # beta in {0,1,2,4}, default beta=2 # 1. Plot the eigenvalues, they lie on the unit circle circular.sample_full_model(size_N=30, haar_mode='Hermite') circular.plot() # 2. Histogram of the angle of more points, should look uniform on [0,2pi] circular.flush_samples() # Flush previous sample circular.sample_full_model(size_N=1000, haar_mode='Hermite') circular.hist()
\(\beta=4\)
Uniform measure i.e. Haar measure on self-dual unitary matrices \(\mathbb{U}\operatorname{Sp}_{2N}\): \(U^{\dagger}U = I_{2N}\)
\[\begin{split} \begin{cases} X \sim \mathcal{N}_{N, M}(0,1) + i~ \mathcal{N}_{N, M}(0,1)\\ Y \sim \mathcal{N}_{N, M}(0,1) + i~ \mathcal{N}_{N, M}(0,1) \end{cases}\\ A = \begin{bmatrix} X & Y \\ -Y^* & X^* \end{bmatrix} \quad A = X+X^{\dagger} = U^{\dagger} \Lambda U\\ eigvals(U).\end{split}\]
See also
Banded matrix model for Circular ensemble
CircularEnsemblein API
Ginibre Ensemble¶
where from the definition in (33) we have \(\left|\Delta(x_1,\dots,x_N)\right| = \prod_{i<j} |x_i - x_j|\).
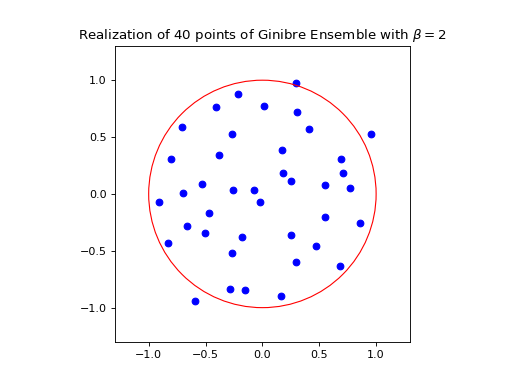
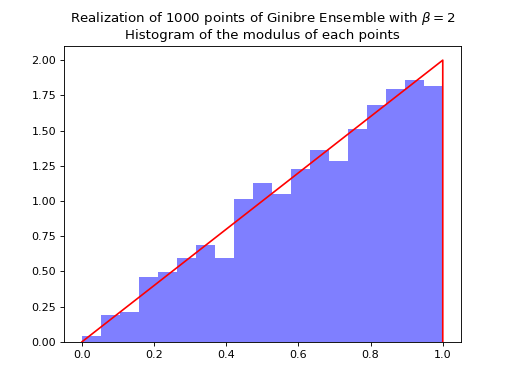
Nomalization \(\sqrt{N}\) to concentrate in the unit circle.
from dppy.beta_ensembles import GinibreEnsemble
ginibre = GinibreEnsemble() # beta must be 2 (default)
ginibre.sample_full_model(size_N=40)
ginibre.plot(normalization=True)
ginibre.sample_full_model(size_N=1000)
ginibre.hist(normalization=True)
See also
GinibreEnsemblein API